Dynamics of infection spread (COVID-19)
Data analysis and modeling techniques from quantitative biology
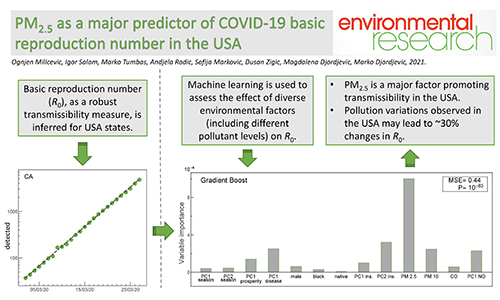
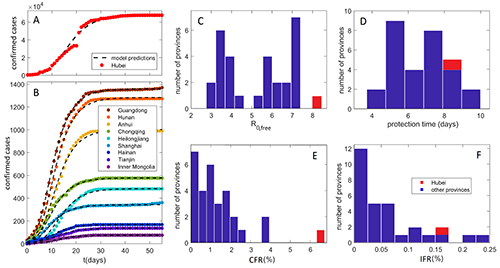
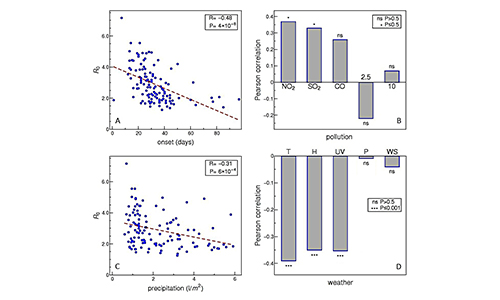
are used to understand COVID-19 transmission and mortality in a population.
The goal is to understand long-term disease behavior if it becomes endemic,
including risk factors for potential future infection outbursts. For examples,
see our COVID-19 work up to now.
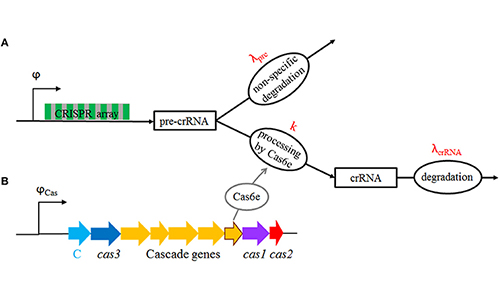
CRISPR/Cas modeling
CRISPR/Cas is an advanced bacterial immune system, which has revolutionized
biotechnology. How is this normally silent system induced? While it is hard to
experimentally observe the system dynamics, this can be more readily done
mathematically, where we use a combination of statistical mechanics and dynamical system
modeling. CRISPR/Cas is a potentially powerful barrier to horizontal gene transfer, so
modeling its regulation also helps to better understand how antibiotic resistance and
virulence genes are disseminated.
CRISPR/Cas bioinformatics
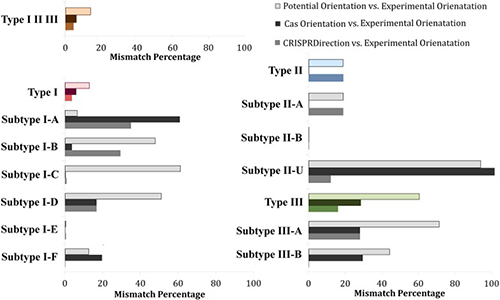
Our research focus is non-canonical CRISPR/Cas functions. It is becoming increasingly
clear that the system is also involved in regulation of endogenous bacterial genes that
are mainly associated with bacterial pathogenicity. As the system is active under poorly
characterized conditions, the target genes can hardly be systematically identified
through experiments alone. We develop bioinformatics methods for
predicting targets of crRNAs and CRISPR-associated small RNAs. We also
computationally work on a related problem or CRISPR/Cas adaptation, specifically what
sequence determinants allow distinguishing self (host) from non-self (viral) DNA.
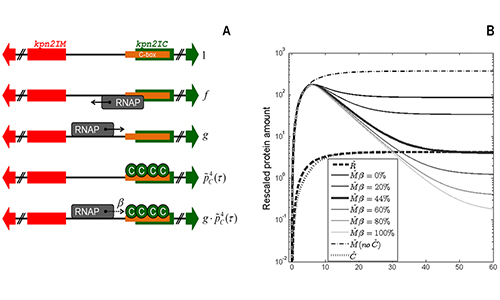
Robustness of bacterial gene circuits to cell growth rate changes
As a model, we use restriction-modification (R-M) systems, which are often spread by
horizontal gene transfer. Consequently, expression of the restriction enzyme and the
methylase has to be tightly regulated during its establishment in a naive bacterial host,
which is often exhibited by specialized transcription regulators.
These systems come with a variety of architectures (convergent, divergent, linear),
where we show that these differences are consistent with few general constraints on the gene
expression regulation. Our current focus is on how these general dynamical features remain robust
under changing cellular growth conditions, that affect crucial intracellular parameters.